The New Biology
A counterintuitive approach to biological design is unlocking life-saving therapies and novel materials
Obvious |

The Humbling Number & Biological Complexity
The humbling number is 10⁶¹⁴. To put that in perspective, your odds of winning the Powerball is on the order of 10⁸. There are 10⁸⁰ total atoms in the universe. To go from 10⁸⁰ atoms to 10⁸¹…takes 10 universes. To go to 10⁸³, 1000 universes. In comparison to those reference points, 10⁶¹⁴ is an unknowably large number.
There are estimated to be 10⁶¹⁴ possible combinations of the human genome.
Biology is the most complex problem space that humans have ever attempted to understand. We should be humbled by the sheer beauty and scale of that complexity, the way that we are when we look at the Grand Canyon or Half Dome (if the Grand Canyon were the depth of near-infinite universes).
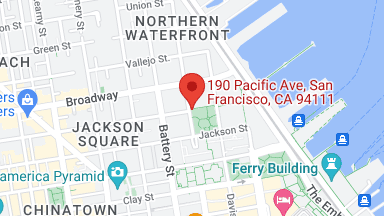
And yet, the primary method of exploring biological function has been to map out the stages of interaction, one step at a time. To deconstruct biological function into discrete components, and create a mental map (see below—sometimes an actual map) to trace the chain of events that explain some observed behavior.
While many therapeutic breakthroughs have come from building out causal biological associations, in many other instances the underlying mechanisms are far too complex and non-deterministic to take this simplistic approach.
As a result, we are now running up against a wall.
Eroom’s Law and Research Productivity
The limitations of this top-down approach to biology have contributed to a massive bottleneck in developing new materials, cures for diseases, and diagnostics.
In material science, the vast majority of materials in the world around us is derived from refining petroleum. This is a process that has been unchanged for over 100 years. Much of the modern progression of materials has actually come from the world of biology, where there is a wide variety of microbes that can produce materials of interest. Even within the smaller biological design space of microbes, which have 4000 genes vs. the 20,000 in humans, much is misunderstood. Many genes within microbes are only partially characterized, and upwards of 25% have no known function of any kind. This incomplete biological map has limited our ability to engineer new microbes and new materials.
In the pharmaceutical world, the limitations of modern drug development is best summarized by Eroom’s Law (reverse Moore’s Law). This is the observation that the inflation-adjusted cost of bringing a new drug to market doubles every nine years.
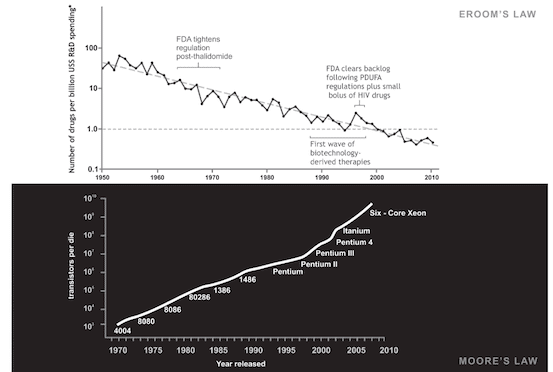
Due to the limitations in top-down biological design, the productivity of new materials, new medical diagnostics, and new treatments is in crisis. We’re not just leaving massive market opportunities on the table. We’re leaving cures, solutions to global challenges, and more on the table.
Don’t (Re) Reinvent the Wheel
The “break down a problem into small, modular components” approach has historically been a helpful shortcut to problem-solving. This has been accomplished by creating a set of building blocks that can loosely be associated with some desired end effect, and assembled together towards a more complex solution. And this alphabetic, rules-driven approach has been effective across a wide range of spaces—but a new methodology is taking over.
As I’ve written about before (The New Intelligence), previous attempts at “alphabetic” solutions across a large number of spaces has been upended by a technique called end-to-end machine learning over the last five years. This has led to breakthrough performance in every one of these fields:
- Speech Recognition (Chomsky’s “Universal Grammar” Symbolics → Deep Learning Models): Power seamless speech-to-text transcription and all modern chatbots like Siri and Alexa.
- Image Recognition (Feature Dictionaries → Convolutional Neural Networks): Enable computers to now have image processing capabilities that exceed human abilities. This applies to general image tagging as well as technical imaging fields like radiology and pathology.
- Scouting in Sports (Scouting + Coaching “Intuition” → Moneyball): Power the drafting and teambuilding decisions across pro-sports.
- Personal Credit (FICO Score → Data-Driven Risk Modeling): Companies like Tala allow for fair access to credit for unbanked populations that don’t have credit scores.
What do all of these approaches have in common? They reject the notion of starting with a causal set of rules that encode the solution to a problem. Instead, a closed feedback loop is created such that a model can be trained over a period of time. Rounds upon rounds of experimentation and testing help to search for the optimal solution.
Given the right conditions to iteratively test and learn, any problem can be turned from a design problem to a search problem.
Take building a bridge as an example.
Design Approach: The “design” approach to building a bridge is to understand the fundamental rules of physics that govern such a structure. Gravity, material properties, stress loads, etc. Then, design a bridge that helps to dissipate the stress so that the force defined by the underlying physics doesn’t overload the bridge. Armed with the correct set of core principles around physics and the sheer willpower of human imagination, an architect like William Howe may be able to come up with a suitable solution.
Search Approach: The “search” approach would be founded on a radically different set of principles. The driving principle of this approach is that it must be possible to build and test bridges quickly, cheaply, and with no real danger associated with failure. If this were true, the search approach would design an experimental cycle that builds a bridge arbitrarily, tests it, learns from the failures, and then incorporates those learnings into a new bridge design. After sufficient cycles of iteration, a winning bridge design would emerge. This bridge may end up with some classic bridge elements like a truss or an arch, but it doesn’t depend on any understanding of physics, or a brilliant mind like Howe’s.
Of course, with the search approach we are talking about machine learning.
ML is essentially this guided trial-and-error process—a method of running many experiments to brute force a solution to a problem. The cheaper and faster you can run an experiment, the faster you can learn.
You see this work well in problems like Go, because the cost of playing Go is nearly $0 and the speed of a “Go experiment” is nearly instantaneous. Without any fundamental Go principles to start, AlphaGo Zero played five million games of Go with itself in three days to iterate on a strategy that allowed it to beat the top player in the world.

Turning Biology Into a Search Problem
Biology is unlike Go, in that we don’t understand all of the rules of biology and can’t simulate biology perfectly. As a consequence, we’re unable to represent biological space purely in simulation the way that Go games can be played digitally. Therefore, experiments in biology have to be compiled and tested in the real world through physical experiments. While we are not quite at Go-levels of experimental speed and cost with biology, we are headed in that direction. A few important forces are coming together to give us a new way of exploring biology at massive scale, which allow researchers to approach challenges in new ways.
Lab Automation & Data Instrumentation
High-throughput lab robotics and multiplexed experimentation methods allow for experiments to run at a scale never achieved before (in some cases hundreds of thousands of experiments at once). Additionally, the most advanced computational biology teams are realizing that not all experimental data is created equally. These teams are serious about machine learning for biology, and they are custom designing and instrumenting experiments with generating rich, consistent, labeled data as the primary objective.
Genomic Datasets
The cost of sequencing, or reading genetic data, is dropping faster than Moore’s Law (it cost $3B to sequence the human genome in 2001, having dropped to ~$1000 today). We can take the empirical readouts from these high-throughput, data-rich lab experiments, and then correlate them to the genetic datasets that come from sequencing. This creates a great set of reference points for machine learning.
Machine Learning
An increasingly mature machine learning toolkit completes the formula. Biological data, genomics, and experimental readouts can feed into sophisticated classifiers that build correlative models and help to isolate commonalities in the high-performing experiments. These are then used to design the next set of experiments. This accelerated evolution will play out over subsequent cycles of experiment, learn, design, build.
All of these factors come together to translate a biological design problem into a data science problem.
We have an incredibly powerful, and proven, modern toolkit at our disposal to attack these biological design challenges in a new way. Most importantly, this new approach to biology allows us to circumvent the traditional first step of mapping out the underlying principles of biological function. Even without a complete a priori plan of some complex biological system, researchers can design experiments and start learning what works.
At Obvious, we’ve been big believers of this computational, experiment-driven approach to biology for years. We have been fortunate to be investors behind some of the most visionary companies that are moving this field forward, and we are seeing the tremendous results from the inside.
Zymergen’s machine learning approach to microbial engineering has led to large industry partnerships and the discovery of completely new materials. For example, developing cellular processes to desalinate water more efficiently, creating biological molecules that will help build more comfortable medical prosthetics, and engineering microbes that could clean up underwater oil spills.
Recursion Pharmaceutical’s computer vision platform for drug discovery has uncovered multiple drugs that the company is now moving into clinical trials. The company approaches drug discovery in a completely new way and gives hope to patients who have diseases that aren’t yet understood.
And as we just announced, LabGenius’ protein engineering follows a closed loop synthetic biology flywheel blending simulation and machine learning (in digital space) and high throughput, in-vitro experimentation (in biological space). This search process allows LabGenius to create proteins never before found in nature and traverse the amino acid space in a methodical and comprehensive way.
To us, this signals the beginning of a golden era in biological research with implications across multiple global, trillion-dollar industries. It is truly amazing to see how quickly advances are being made in machine learning, experiment instrumentation, and genetic sequencing/synthesis.
However, many of these ideas are the antithesis of first-principles approach to biology that scientists are trained on. Predictably, there has been a great deal of pushback from prominent researchers, veterans from big pharma, and investors. The open question is whether we can shift our mindsets as quickly as the technology is developing. I believe that we can.